Resource Information

유전자의 영향으로 무핵과(seedless)가 되는 힘로드(Himrod) 품종과 GA 처리를 통해 무핵과를 유도하는 탐나라(Tamnara) 품종 간에 개화 전후의 유전자 발현 양상을 분석하여 공통점과 차이점을 살펴보고자 함. |
실험계획
DEG 분석 / Control vs. Treatment
Control
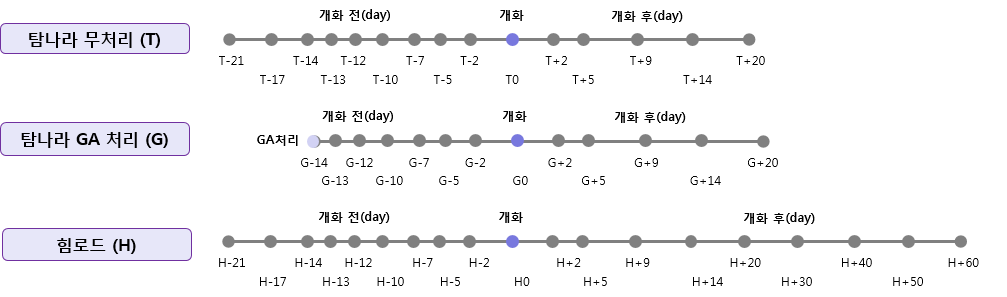
1.탐나라 무처리 (T)
2.탐나라 무처리 (T)
3.힘로드 (H)
2.탐나라 무처리 (T)
3.힘로드 (H)
VS
Treatment
1.탐나라 무처리 (T)
2.탐나라 GA 처리 (G)
3.힘로드 (H)
2.탐나라 GA 처리 (G)
3.힘로드 (H)
KEGG metabolic pathway & GC/MS
Metabolism
-
Carbohydrate Metabolism
-
00010 Glycolysis / Gluconeogenesis

00020 Citrate cycle (TCA cycle)

00030 Pentose phosphate pathway

00040 Pentose and glucuronate interconversions

00051 Fructose and mannose metabolism

00052 Galactose metabolism

00053 Ascorbate and aldarate metabolism

00500 Starch and sucrose metabolism

00520 Amino sugar and nucleotide sugar metabolism

00620 Pyruvate metabolism

00630 Glyoxylate and dicarboxylate metabolism

00640 Propanoate metabolism

00650 Butanoate metabolism

00660 C5-Branched dibasic acid metabolism

00562 Inositol phosphate metabolism

-
00190 Oxidative phosphorylation

00195 Photosynthesis

00196 Photosynthesis - antenna proteins

00710 Carbon fixation in photosynthetic organisms

00910 Nitrogen metabolism

00920 Sulfur metabolism

-
00061 Fatty acid biosynthesis

00062 Fatty acid elongation

00071 Fatty acid degradation

00072 Synthesis and degradation of ketone bodies

00073 Cutin, suberine and wax biosynthesis

00100 Steroid biosynthesis

00561 Glycerolipid metabolism

00564 Glycerophospholipid metabolism

00565 Ether lipid metabolism

00600 Sphingolipid metabolism

00590 Arachidonic acid metabolism

00591 Linoleic acid metabolism

00592 alpha-Linolenic acid metabolism

01040 Biosynthesis of unsaturated fatty acids

-
00230 Purine metabolism

00240 Pyrimidine metabolism

-
00250 Alanine, aspartate and glutamate metabolism

00260 Glycine, serine and threonine metabolism

00270 Cysteine and methionine metabolism

00280 Valine, leucine and isoleucine degradation

00290 Valine, leucine and isoleucine biosynthesis

00300 Lysine biosynthesis

00310 Lysine degradation

00330 Arginine and proline metabolism

00340 Histidine metabolism

00350 Tyrosine metabolism

00360 Phenylalanine metabolism

00380 Tryptophan metabolism

00400 Phenylalanine, tyrosine and tryptophan biosynthesis

-
00410 beta-Alanine metabolism

00430 Taurine and hypotaurine metabolism

00450 Selenocompound metabolism

00460 Cyanoamino acid metabolism

00480 Glutathione metabolism

-
00510 N-Glycan biosynthesis

00514 Other types of O-glycan biosynthesis

00531 Glycosaminoglycan degradation

00563 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis

00603 Glycosphingolipid biosynthesis - globo series

00604 Glycosphingolipid biosynthesis - ganglio series

00511 Other glycan degradation

-
00730 Thiamine metabolism

00740 Riboflavin metabolism

00750 Vitamin B6 metabolism

00760 Nicotinate and nicotinamide metabolism

00770 Pantothenate and CoA biosynthesis

00780 Biotin metabolism

00785 Lipoic acid metabolism

00790 Folate biosynthesis

00670 One carbon pool by folate

00860 Porphyrin and chlorophyll metabolism

00130 Ubiquinone and other terpenoid-quinone biosynthesis

-
00900 Terpenoid backbone biosynthesis

00902 Monoterpenoid biosynthesis

00909 Sesquiterpenoid and triterpenoid biosynthesis

00904 Diterpenoid biosynthesis

00906 Carotenoid biosynthesis

00905 Brassinosteroid biosynthesis

00908 Zeatin biosynthesis

00903 Limonene and pinene degradation

-
00940 Phenylpropanoid biosynthesis

00945 Stilbenoid, diarylheptanoid and gingerol biosynthesis

00941 Flavonoid biosynthesis

00944 Flavone and flavonol biosynthesis

00942 Anthocyanin biosynthesis

00950 Isoquinoline alkaloid biosynthesis

00960 Tropane, piperidine and pyridine alkaloid biosynthesis

00232 Caffeine metabolism

00965 Betalain biosynthesis

00966 Glucosinolate biosynthesis

Genetic Information Processing
-
Transcription
-
03020 RNA polymerase

03022 Basal transcription factors

03040 Spliceosome

-
03010 Ribosome

00970 Aminoacyl-tRNA biosynthesis

03013 RNA transport

03015 mRNA surveillance pathway

03008 Ribosome biogenesis in eukaryotes

-
03060 Protein export

04141 Protein processing in endoplasmic reticulum

04130 SNARE interactions in vesicular transport

04120 Ubiquitin mediated proteolysis

04122 Sulfur relay system

03050 Proteasome

03018 RNA degradation

-
03030 DNA replication

03410 Base excision repair

03420 Nucleotide excision repair

03430 Mismatch repair

03440 Homologous recombination

03450 Non-homologous end-joining

Environmental Information Processing
-
Membrane Transport
-
02010 ABC transporters

Cellular Processes
-
Transport and Catabolism
-
04144 Endocytosis

04145 Phagosome

04146 Peroxisome

04140 Regulation of autophagy

Organismal Systems
-
Environmental Adaptation
-
04712 Circadian rhythm - plant

04626 Plant-pathogen interaction




